How can digitalisation support interactions between tissue biobanks, and why is this important? The BRoTHER project may have the answers.
The project BRoTHER (Biobank Research on Telemedical Approaches for Human Biobanks in a European Region) was created to analyse the burdens that should be overcome when clinical-related biobanks, especially tissue biobanks, want to co-operate. An important issue within networking between different biobanks in the clinical context is digitalisation. Digitalisation is of great value in sharing information in an appropriate manner. However, interactions of co-operating biobanks, especially in different countries, are facing further challenges regarding the exchange of knowledge and data.
Towards a digital pathology framework
Part of the project is to create a digital pathology framework which enables an easy exchange of image files and makes it simple to perform joint research projects (Fig. 1). Thus, it is possible to check already in the planning phase of a project whether the appropriate quality and number of suitable biobank samples are available at one of the partner tissue biobanks. If a second consultation on histopathological data is required in a project, a simple exchange between the participating project partners is possible.
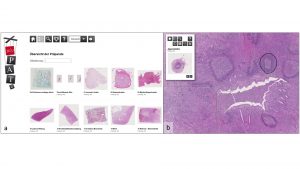
As a basis for the data exchange, whole-slide imaging (WSI) and virtual microscopy (VM) technologies are used, which were developed as a user-friendly e-learning tool for teaching in histopathology.1,2,3 After transforming the Pate platform in a bilingual tool, it will serve as a prototype for a digital pathology framework as an interactive reference pathology tool. An optimisation for desktop applications and smaller screens was implemented, and several feedback loops during development resulted in a smart system for easy-to-use online consultations and image exchanges.
The relevance and accuracy of telemedical approaches in pathology has been demonstrated not only in the workflow of histopathological diagnostics but also about 3D microscopy.4,5 Both clinical and research-based applications require secure access and handling of the image files. The use of sophisticated WSI and VM technologies in a regional network of tissue biobanks not only provides access to expertise but also facilitates new research projects.
Histopathological report data
A second topic within digitalisation is the extraction of relevant data from histopathological reports, which contain relevant information for the sample such as the exact anatomical location of the sample and the tumour dimensions. Furthermore, specific information, such as necrosis, inflammation, fibrosis or wound-healing reactions, are noted in the histopathological report. Of special value are reports on malignant tumours.
These contain not only statements on staging and grading but also for the expression of certain molecules. These in turn can serve as specific target points in personalised medicine. The aim of such new therapies is to attack the tumour more effectively, with fewer side effects at the same time.6
Especially in children, long-term remissions can be achieved with conventional therapies in 80% of the cases. Nevertheless, malignant tumours are still one of the major causes of morbidity and mortality in children and young adults which should be further reduced.7
Histopathological reports can provide information on the cellular properties, such as tumour vascularisation, proliferation behaviour of the tumour cells or immunological reactions of the surrounding tissue, as well as aggressiveness and invasiveness. Extracting this relevant information from a histopathological report is needed on the one hand for the understanding of tumour biology but also for the development of new therapeutic options.
Therefore, a minimal histopathological information-set for biobank samples will be developed. This dataset, which is linked to the biobank sample and stored digitally, should serve to better assess its potential suitability for certain research projects. One way to achieve this goal would be to use formalised histopathological reports.
Currently, the reality consists of mainly unstructured continuous text. From the point of view of the pathologist, however, this text makes it easier to express subtle but thoroughly relevant information and its relationships with one another. Nevertheless, every pathology report ends with a specific epicrisis. The epicrisis clearly defines the anatomical localisation of the sample, the grading and all other relevant information on possible therapeutic approaches.
Thus, the epicrisis can be seen as a summary of all the essential points of a histopathological assessment. Although one could therefore use the epicrisis to extract the desired data, the text still contains more information. The development of personalised medicine is based on the exact description of tumours and their characteristics. Automated data extraction from formalised texts or structured tables using digital tools is easy because it is more of a thesaurus than a text. However, extraction from a continuous text poses a great challenge.
To address this issue, we compare pathology reports from tumour patients within the BRoTHER network to each other’s structure and formalisation levels. Here, the above-mentioned student exchange programme plays an important role.
Graduate and postgraduate students of one institute visit a partner institute and learn how their pathological reports are built up. Based on these visits, formalised structure-organograms are created for pathology reports of each institute. These in turn will then be compared with each other in workshops.
Minimal information sets
Another task of the students is to evaluate so-called ‘minimal information sets’ for each laboratory. During their visit, the students evaluate what data from a pathological report is needed for each biobank. These sets are then also discussed at workshops and implemented in the partner’s biobank concept. Thus, concepts are permanently developed further on how to obtain acceptable data from the pathology reports for all partners and how they can be used for the biobank network, as proposed by the System of BBMRI-ERIC.8
References
- Development of Pat-e an innovative e-learning framework to teach histopathological features. MAICUM grant program, University Medical Centre Mainz (2011-2013; Christoph Brochhausen-Delius)
- Development of a quiz-mode for Pate. MAICUM grant program, University Medical Centre Mainz (2014-2015; Christoph Brochhausen-Delius)
- Telemedicine in the Euroregion POMERANIA-POMERANIA-Netzwerk (EU-EFRE-program; ref-nr: INT-08-0001; Matthias Evert)
- Kalinski T, Zwönitzer R, Sel S, Evert M, Günther T, Hofmann H, Bernarding J, Roessner A (2008) Virtual 3D microscopy using multiplane whole slide images in diagnostic pathology. Am J Clin Pathol 130: 259-264
- Ribback S, Flessa S, Gromoll-Bergmann K, Evert M, Dombrowski F (2014) Virtual slide telepathology with scanner systems for intraoperative frozen-section consultation. Pathol Res Pract. 210: 377-382
- Klement GL, Arkun K, Valik D, Roffidal T, Hashemi A, Klement C, Carmassi P, Rietman E, Slaby O, Mazanek P, Mudry P, Kovacs G, Kiss C, Norga K, Konstantinov D, André N, Slavc I, van Den Berg H, Kolenova A, Kren L, Tuma J, Skotakova J, Sterba J (2016) Future paradigms for precision oncology. Oncotarget 7: 46813-46831
- Steˇrba J, Steˇrbová S, Kodytková D, Valík D, Demlová R (2014) Clinical evaluation of new drugs against orphan diseases in oncology – the current situation in Europe and in our country. Vnitr Lek 60 Suppl 2: 80-5
- Holub P, Greplova K, Knoflickova D, Nenutil R, Valik D (2012) The biobanking research infrastructure BBMRI_CZ: a critical tool to enhance translational cancer research. Klin Onkol 25: 2S 78-81
Christoph Brochhausen
Institute of Pathology
+49 941 944 66 36
christoph.brochhausen@klinik.uni-regensburg.de
Please note, this article will appear in issue 9 of Health Europa Quarterly, which is available to read now.