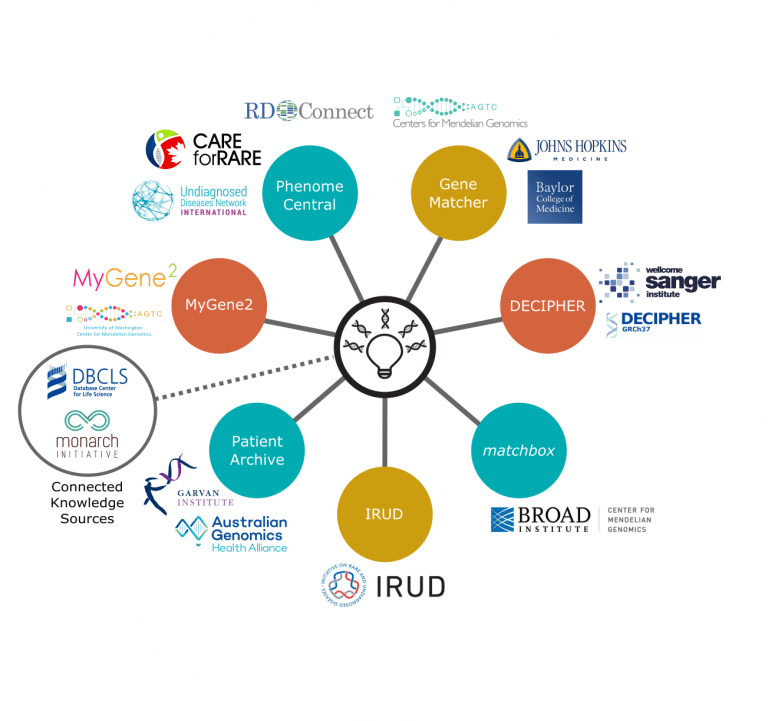
The Matchmaker Exchange (MME) was initiated in October 2013 as a community-organized effort to connect clinicians and researchers with rare cases. Matchmakers had evolved in several national and international efforts to identify new Mendelian disease genes, such as:
- FORGE and Care4Rare Canada, which developed PhenomeCentral (phenomecentral.org);
- The Centers for Mendelian Genomics (National Human Genome Research Institute), which developed GeneMatcher (genematcher.org); and
- The Wellcome Sanger Institute, which developed the DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources (DECIPHER) project in the UK (decipher.sanger.ac.uk).
The goal of the MME was to develop a connection (application programming interface; API) that would allow exchange of information across these and other matchmakers so that cases would not have to be deposited in multiple databases.
This API was launched as a federated system in July 2015. A special issue of Human Mutation in October 2015 described the MME, its API, connected matchmakers, and a few examples of successful disease gene discovery. The MME is a driver project of both the Global Alliance for Genomics and Health (GA4GH, ga4gh.org) and the International Rare Disease Research Consortium (IRDiRC.org).
Since its initial launch, four additional connected nodes have joined the federated network:
- Initiative on Rare and Undiagnosed Disorders (IRUD), Japan;
- matchbox, US;
- MyGene2, US; and
- Australian Genomics (Patient Archive), Australia.
It is easy to submit cases to a matchmaker, and guidance regarding which matchmaker to use is available at http://www.matchmakerexchange.org/i_am_a_clinician_laboratory.html.
There is also extensive guidance about joining as an entire database on the MME website, as well as monthly calls of MME participants which are open to all. The MME is governed by a steering committee (SC) comprised of one representative from each connected matchmaker and a one from the GA4GH and IRDiRC. Most decisions are made on the monthly calls of all MME participants, but if necessary between calls or in cases of disagreement, the SC votes.
The MME has periodic in-person working meetings to advance the platform in support of rare disease gene discovery. In addition, there is a meeting as part of the annual GA4GH meeting to align the MME with other genomic data-sharing initiatives.
Information exchanged
Only cases submitted by researchers and/or clinicians can be exchanged over the MME. Patient-initiated connections are feasible within MyGene2 and GeneMatcher, but are not yet permissible over the MME. The API requires that the submitter be registered in one node, a submission ID, and candidate gene information (such as an Entrez or Ensembl Gene ID); optional additional information includes disorder number (OMIM or Orphanet ID), and phenotypic features exchanged as Human Phenotype Ontology (HPO) IDs (although GeneMatcher will also accept PhenoDB IDs and ICHPT IDs). There is an end-user agreement required of any user of the MME:
- To make no attempt to identify individual patients in any MME database;
- To enable all cases submitted for querying to be stored in the query-initiating database for future matching;
- To obtain permission from the source of the matching data before publishing or presenting the results of queries, and to respect the privacy of patients in any publications and presentations;
- To acknowledge the MME, and the specific MME services that supported any discoveries in publications, as appropriate. See http://www.matchmakerexchange.org/citing;
- To acknowledge that all activities and requests for matches will be logged. Logging information will include full name, email address, time stamp of activity, query data, and the response; and
- To acknowledge that logging information will be available to the originating and the queried databases but it will not be shared outside MME. Logging information will not be used to gain knowledge of user’s research activities for competitive purposes. It will be used to monitor MME services and to generate aggregate statistics to report on performance (e.g. at public events such as conferences).
Matchmakers are required to log and track queries and authenticate submitters. To become a bona fide member of the MME any new matchmaker must establish connections to at least two other matchmakers.
Matchmaker Exchange’s two-tier consent requirements
Submissions involving only candidate genes and HPO terms do not require explicit informed consent, as this can be considered an extension of clinical care to establish a diagnosis. Submissions from a research context or including detailed phenotypic descriptions or multiple variants that could allow identification require informed consent. The onus for obtaining and assuring consent rests with the submitter. A detailed description of the consent structure is available at matchmakerexchange.org under the Resources tab.
Connected nodes
GeneMatcher was developed by the Baylor-Hopkins Center for Mendelian Genomics to enable connections among researchers and clinicians with an interest in the same gene. It has been expanded to include patient-deposited cases as well as phenotypes and disorders, although a candidate gene is necessary for matching. GeneMatcher accepts submissions directly from any instance of PhenoDB, as well as direct submissions. Currently, GeneMatcher contains over 22,700 submissions from 5,372 users of over 9,100 genes, 49% of which have matched, some across the MME.
PhenomeCentral was developed by the Hospital for Sick Children and University of Toronto, Canada. PhenomeCentral uses an interface based on the PhenoTips program and allows for granular consent-based controls of fields entered for any patient, including phenotypes, lists of candidate genes, or a whole exome Variant Call Format (VCF) file. Users are also able to share data directly from their local PhenoTips implementation.
PhenomeCentral has over 2,500 phenotyped cases from 952 users, and has enabled over 1,100 matches across the Matchmaker Exchange. PhenomeCentral is the matchmaker used by the Care4Rare Canada project, the Undiagnosed Disease Network in the US, and Undiagnosed Disease Networks International.
DECIPHER is the oldest and largest matchmaker with over 26,000 open-access patient records and more than 14 years’ experience. All data available for query through the MME is open-access and requests for connection go through a database administrator. DECIPHER includes single-nucleotide variants, copy number variants, and HPO annotations for submitted cases.
MyGene2 has been accepting patient-initiated case-level data deposition since April 2016 and currently contains 1,576 family profiles that are completely open-access. It has been connected to the MME since summer 2016. Only cases attached to a clinician or researcher that include candidate novel Mendelian disease genes can be submitted for querying across the MME.
Broad matchbox is the MME node at the Broad Center for Mendelian Genomics (Broad CMG). The software system that powers this node (also named “matchbox”) was developed as a collaboration between the Broad CMG and the Monarch Initiative.
At the Broad CMG, matchbox is embedded inside the seqr application, a rare disease research platform developed by the MacArthur Laboratory. seqr allows the collaborative analysis of genomic data and currently contains over seventeen thousand patients. matchbox is built distinctly from seqr and is portable, enabling other genomic centres to add it to their existing analysis platforms and connect to the MME as easily as possible. matchbox is freely available as open-source software under a BSD Licence.
AGHA Patient Archive The Australian Genomics Patient Archive was connected to the MME in 2017. It includes unsolved cases that have undergone exome or genome sequencing as part of the Australian national effort.
IRUD The Japan Agency for Medical Research and Development (AMED)-led program Integrating Knowledge for Diagnoses (IRUD) is combining expertise and technology to develop a systematic approach to supporting patients with medically unidentified conditions. IRUD operates “IRUD Exchange” which is based on Patient Archive. The platform is available for access only to clinicians and researchers registered with the national initiative.
IRUD joined MME in December 2017 and currently consists of over 1,200 cases.
Connected knowledge sources
Monarch Initiative enables the identification of animal models for human diseases based upon features. Phenotypes in human patients and animal models can be computationally compared and evaluated despite being recorded using different vocabularies. These types of comparisons can aid identification of causative variants during exome analysis, even when human mutations may not have been previously identified.
The Monarch Initiative accepts queries across the MME and returns possible animal models. It does not initiate queries across the MME.
PubCaseFinder is a diagnosis support system for rare diseases supported by the Database Center for Life Sciences (Japan), which contains more than one million case reports harvested from PubMed that have been annotated for concepts of rare diseases and phenotypes.
Future plans
The MME hopes to expand to include other matchmakers, particularly from areas of the world that are currently under-represented. The MME technical group is working on developing both one-sided hypothesis matching (a candidate gene against genome-wide variant data with no flagged candidate genes) and hypothesis-free matching (unsolved case against unsolved case with phenotypic and genome-wide variant data).