Semra Ince and Christian Herrmann of Ruhr-Universität Bochum, Germany, discuss the role of oligomerisation in proteins.
Throughout known life there are 20 standard amino acids and a genetic code, which unambiguously dictates the chain of polypeptide built from those amino acids. It is the particular chemistry of each amino acid and their order within the chain which in turn makes the chain arrange into a three-dimensional construct, the protein – and we now can delve further into proteins, looking specifically at oligomerisation. The so-called ‘workhorse’ within the organism takes a variety of roles: enzymes, ion channels, hormones, antibodies and structural proteins are only a few examples of protein classes.
Oligomerisation as a regulatory mechanism
This is how the story of life begins and rapidly becomes complicated when considering that each cell harbours thousands of different proteins which need to be orchestrated such that they can fulfil their particular task at the right place and at the right time.1 Herein, oligomerisation represents a potent regulatory mechanism to ensure the proper functioning of proteins. Oligomerisation is the assembly of protein units into macromolecular complexes through specific interactions with either the same type of protein (homo-oligomers) and/or other types of proteins (hetero-oligomers). While only one third of all proteins subsist as monomers, interestingly, the vast majority of proteins gain their functionality only after assembly into oligomeric complexes which only emphasises the regulatory principle of oligomerisation. In fact, this only gets more pronounced when considering that many proteins execute different activities in the monomeric state as compared to the oligomeric state.2,3
It is easy to comprehend that from an evolutionary point of view it is more reasonable to obtain multiple copies of one protein from a shorter gene to form an oligomer rather than obtaining a single polypeptide chain with multiple repeats from a larger sized gene; a scenario that negatively correlates with the foldability of the protein. The efficiency of oligomerisation can be best demonstrated by viral particles where a minimum size of a genome serves for multiple copies of only a few proteins to constitute an entire viral capsid. Moreover, the assembly of those proteins through specific sites leads to a symmetry which improves the stability of the particle. Still maintaining stability, it is not necessarily that the oligomeric complex is of static nature. Rather, dynamic assembly and disassembly is often seen in Nature, for instance at cytoskeletal proteins which in this manner can ensure not only temporal stability but also plastic flexibility of a cell.
The building of oligomeric structures
The dynamin superfamily of large GTPases represents a group of proteins which accomplish essential cellular functions such as membrane fission and fusion events as well as innate immune functions by their ability to form homo-oligomers. Having a typical multi-domain architecture, these proteins exploit certain sets of subdomains to build dimeric, tetrameric as well as higher oligomeric structures, e.g. spirals, which wrapped around budding vesicles are capable to mediate their fission, and rings, which supply multiple interaction sites for the stable binding of viral proteins, thereby preventing viral propagation.4,5
Human guanylate-binding proteins with seven isoforms comprise a group within the dynamin superfamily with at least five isoforms (hGBP-1 to hGBP-5) shown to share the common feature of nucleotide-dependent oligomerisation. Qualitative cell studies moreover demonstrate that the oligomerisation of the hGBPs is not only restricted to homotypic interactions but also to heterotypic interactions among the hGBP with the consequence of subcellular translocation to compartments being specific to each complex.6 These findings in view of the marginal molecular details, (see our previous article in PEN Health 2), suggest an important contribution of the hGBPs’ capability to oligomerise.
Thus, the characterisation of the hGBPs oligomers by means of biochemical and biophysical techniques using recombinant proteins in a cell-free system is a major topic of our research which, in detail, addresses:
- Quantities of oligomeric species of hGBPs in different nucleotide-bound states
- The dynamics of the oligomers’ assembly and disassembly, especially with the rates of disassembly giving information about the life-time of oligomers
- Affinities of the different oligomers, giving information about their relevance at cellular concentrations
- Subdomains and interfaces being involved in oligomer formation and
- Both requirements for and consequences of oligomerisation with respect to conformational/structural changes and the GTPase activity of each protein.
The self-regulatory activity of hGBP-1
Most of our studies in recent years have focused on the homo-oligomerisation of hGBP-1 which is the only member with a solved crystal structure and plays a central role in many of the immune responses reported for hGBPs. Our findings suggest a highly self-regulatory activity of hGBP-1 linked to binding of its substrate GTP and its hydrolysis in two successive steps.
These events are accomplished through a complex interplay between structural changes and the co-ordination of two subdomains which build homo-dimers from monomers; herein, interactions provided by the catalytic domains serve to stabilise the active sites which substantially enhances the rate of GTP hydrolysis. Interactions between the C-terminal domains on the other hand are controlled by the first GTP hydrolysis step and enhance the lifetime of the dimers, thereby allowing the second hydrolysis step to occur.
These data altogether demonstrate the importance of hGBP-1 dimers as catalytic active modules which on the one hand control the hydrolysis rate of GTP and on the other hand dissociate into inactive units after GTP is consumed.7,8 and unpublished data
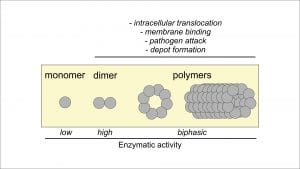
Beside the dimers, more recently we identified even higher oligomeric species of hGBP-1 which we termed polymers; these are built from several thousands of molecules which have disc-like and tubular geometries and provide somewhat altered enzymatic activity as compared to the dimers (Fig. 2).9 Interestingly, these assemblies are only observed for the post-translationally modified form of hGBP-1 which at the C-terminus is equipped with a hydrophobic farnesyl moiety. In this form, hGBP-1 also gains the ability to interact with membranes which reveals as a mechanism competitive to polymerisation. Whether membrane binding of hGBP-1 relies on a certain oligomeric state or if those polymers have a more particular role than the speculated depot function is yet to be fully understood.
The roles of the remaining members (both in the post-translationally modified and non-modified form), which we recently started to investigate, also need to be considered. Our newest data, for instance, show that hGBP-1 also forms hetero-dimers with other hGBP members, like with hGBP-5.8 Interestingly, these hetero-dimers represent similar catalytic competent units which moreover have a similar affinity as the homo-dimers of hGBP-1. Thus, in this case we can assume that each type of dimer is equally likely to occur which, for the first time, unravels a competitive situation between the hGBPs under cellular conditions.
Whether this applies to the remaining members, and how exactly they engage with the hGBP networking based on the exchange of interaction partners, oligomeric species and the subcellular forms part of our ongoing research. Nevertheless, the example of hGBP-1 is evident to demonstrate that protein dimerisation and oligomerisation is most important for the regulation of the enzymatic activity and for defining the subcellular localisation. While for a long time the function of hGBP-1 and its molecular mechanism was sought on the level of the isolated, monomeric enzyme the key for full understanding was found in the formation of dimers and oligomers of this enzyme.
References
- Milo et al. (2010). Nucl Acids Res 38 (suppl 1):D750-D753.
- Marianayagam et al. (2004). The power of two: protein dimerization in biology. Trends Biochem Sci 29(11): 618-25.
- Lesieur (2014). The Assembly of Protein Oligomers – Old Stories and New Perspectives with Graph Theory Chemistry. Chapter 11 in Oligomerization of Chemical and Biological Compounds; ISBN 978-953-51-1617-2.
- Praefcke (2017). Regulation of innate immune functions by guanylate-binding proteins. Int J Med Microbiol pii: S1438-4221(17): 30425-3.
- Faelber et al. (2013). Oligomerization of dynamin superfamily proteins in health and disease. Prog Mol Biol Transl Sci. 117: 411-43.
- Britzen-Laurent et al. (2010). Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5(12): e14246.
- Ince et al. (2017). The human guanylate-binding proteins hGBP-1 and hGBP-5 cycle between monomers and dimers only. FEBS J 284(14): 2284-2301.
- Kutsch et al. (2018; Epub ahead of print). Homo and hetero dimerisation of the human guanylate-binding proteins hGBP-1 and hGBP-5 characterised by affinities and kinetics. FEBS J; doi: 10.1111/febs.14459.
- Shydlovskyi et al. (2017). Nucleotide-dependent farnesyl switch orchestrates polymerization and membrane binding of human guanylate-binding protein 1. Proc Natl Acad Sci U S A 114(28): E5559-E5568.
Semra Ince and Christian Herrmann
Physical Chemistry 1
Ruhr-Universität Bochum
+49 (0)234 32 24173
chr.herrmann@rub.de
http://www.ruhr-uni-bochum.de/proin
This article will appear in issue 7 of Health Europa Quarterly, which will be published in November 2018.