A research team from the Swiss Federal Institute of Technology Lausanne (SFITL) has discovered the two DNA defence systems responsible for the aversion of another cholera pandemic.
Two DNA defence systems protect the bacterial strains from potentially harmful genetic material and viruses amid the ongoing seventh cholera pandemic. This study also demonstrates that the defence systems may have been key in the evolution and success of these strains.
This study has been published in the journal Nature.
What is cholera?
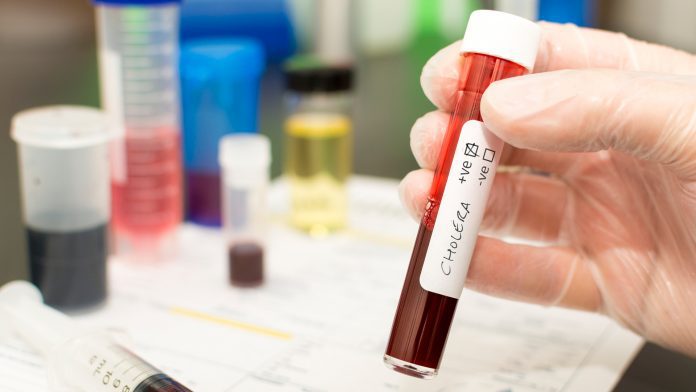
Cholera is caused by the bacterium Vibrio cholerae, which is a waterborne pathogen that infects the gut of humans through contaminated water and food. When ingested, V. cholerae colonises the gut’s inner surface, causing watery diarrhoea that, if left untreated, can lead to severe dehydration and death.
Cholera is still a problem, especially in less-developed or crisis-hit regions. The World Health Organization (WHO) reported that the ongoing seventh cholera pandemic is responsible for up to four million infections, and up to 143,000 deaths each year.
Only a few strains of V. cholerae can cause this cholera pandemic disease, with most being harmless aquatic organisms. This is because the pandemic strains have acquired specialised ‘toolboxes’ of genes and other genetic elements called ‘pathogenicity islands,’ which can turn the bacterium into a pathogen.
What is horizontal gene transfer?
Strains that cause a cholera pandemic have acquired pathogenicity islands through a process known as ‘horizontal gene transfer.’ This refers to bacteria that share genes both within and across species. Horizontal gene transfer is a powerful driver of bacterial evolution because it can quickly endow bacteria with novel abilities that help them adapt and survive. But it is also indiscriminate, passing on genes that are unnecessary or even harmful to their new host.
Horizontal gene transfer often involves plasmids, which are self-replicating circular pieces of DNA located in bacteria that can carry up to hundreds of genes. But strains of V. cholerae that are currently causing the ongoing seventh cholera pandemic rarely carry plasmids, while plasmids are abundant in related strains isolated from the environment instead of patients.
This surprising phenomenon caught the attention of scientists at EPFL, who decided to investigate: “We wanted to find out why plasmids are so rare in the seventh cholera pandemic clade of V. cholerae, shedding light on how bacterial pathogens evolve,” explained Professor Melanie Blokesch, who led the study with Milena Jaskólska and David W. Adams at EPFL’s School of Life Sciences.
How did this impact the cholera pandemic?
First, the researchers introduced a small model plasmid into V. cholerae strains from the sixth and seventh cholera pandemics, as well as non-pandemic strains isolated from different water bodies. They then tracked the plasmid’s stability over the course of a variety of generations. Surprisingly, the model plasmid persisted in all strains, but was quickly eliminated from the seventh pandemic ones.
Following this, scientists utilised genetic engineering methods to identify the parts of the V. cholerae genome responsible for this loss. This approach led to the discovery of two novel defence systems that work together to eliminate plasmids and are encoded within two distinct pathogenicity islands.
What did the team discover about these defence systems?
Researchers named the systems ‘DNA defence modules’ (Ddm). The first – DdmDE – is constructed of two proteins that target and degrade small plasmids in a process helped by a second defence system – DdmABC.
It was revealed that the second system has a much broader role in bacterial defence. Not only can it enhance the elimination of small plasmids, but it can also turn against the host cell, thus degrading its DNA and triggering a form of cell suicide. Essentially, DdmABC protects the bacterial population against viruses by killing infected cells before the virus has time to replicate and spread.
Additionally, the team also discovered that DdmABC targets large plasmids that often carry huge arrays of antibiotic-resistance genes that can persist by jumping from one bacterium to the next, spreading multidrug resistance.
“This finding might explain why the recent pandemic strains mainly carry antibiotic resistance integrated in their genome and not on plasmids,” noted Blokesch.
“The combined activity of these two defence systems solves the long-standing mystery of the missing plasmids in the seventh cholera pandemic V. cholerae strains,” concluded the researchers. “Furthermore, our discovery suggests that the ability of the seventh pandemic strains to defend against mobile genetic elements has likely played a key role in their evolution and success.”
Funding from this project was provided by:
- The European Research Council (ERC);
- The Swiss National Research Foundation (SNSF) EPFL; and
- The Howard Hughes Medical Institute (HHMI).